Comparing linear and Additive logistic regression
Library setup
Read in the data
| X | purchase | n_acts | bal_crdt_ratio | avg_prem_balance | retail_crdt_ratio | avg_fin_balance | mortgage_age | cred_limit |
|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 11 | 0.000 | 2494.414 | 0.000 | 1767.197 | 182.00 | 12500 |
| 2 | 0 | 0 | 36.095 | 2494.414 | 11.491 | 1767.197 | 138.96 | 0 |
| 3 | 0 | 6 | 17.600 | 2494.414 | 0.000 | 0.000 | 138.96 | 0 |
| 4 | 0 | 8 | 12.500 | 2494.414 | 0.800 | 1021.000 | 138.96 | 0 |
| 5 | 0 | 8 | 59.100 | 2494.414 | 20.800 | 797.000 | 93.00 | 0 |
| 6 | 0 | 1 | 90.100 | 2494.414 | 11.491 | 4953.000 | 138.96 | 0 |
clean up the data
look at the distribution of the data
csale_clean |>
select_if(is.numeric) |>
gather() |>
ggplot(aes(value)) +
geom_density(adjust = 3, col = "#CC9933", fill = "firebrick", alpha = 0.5) +
facet_wrap(~key,scales="free")+
theme(axis.text.x = element_text(angle = 60))+
theme_tufte()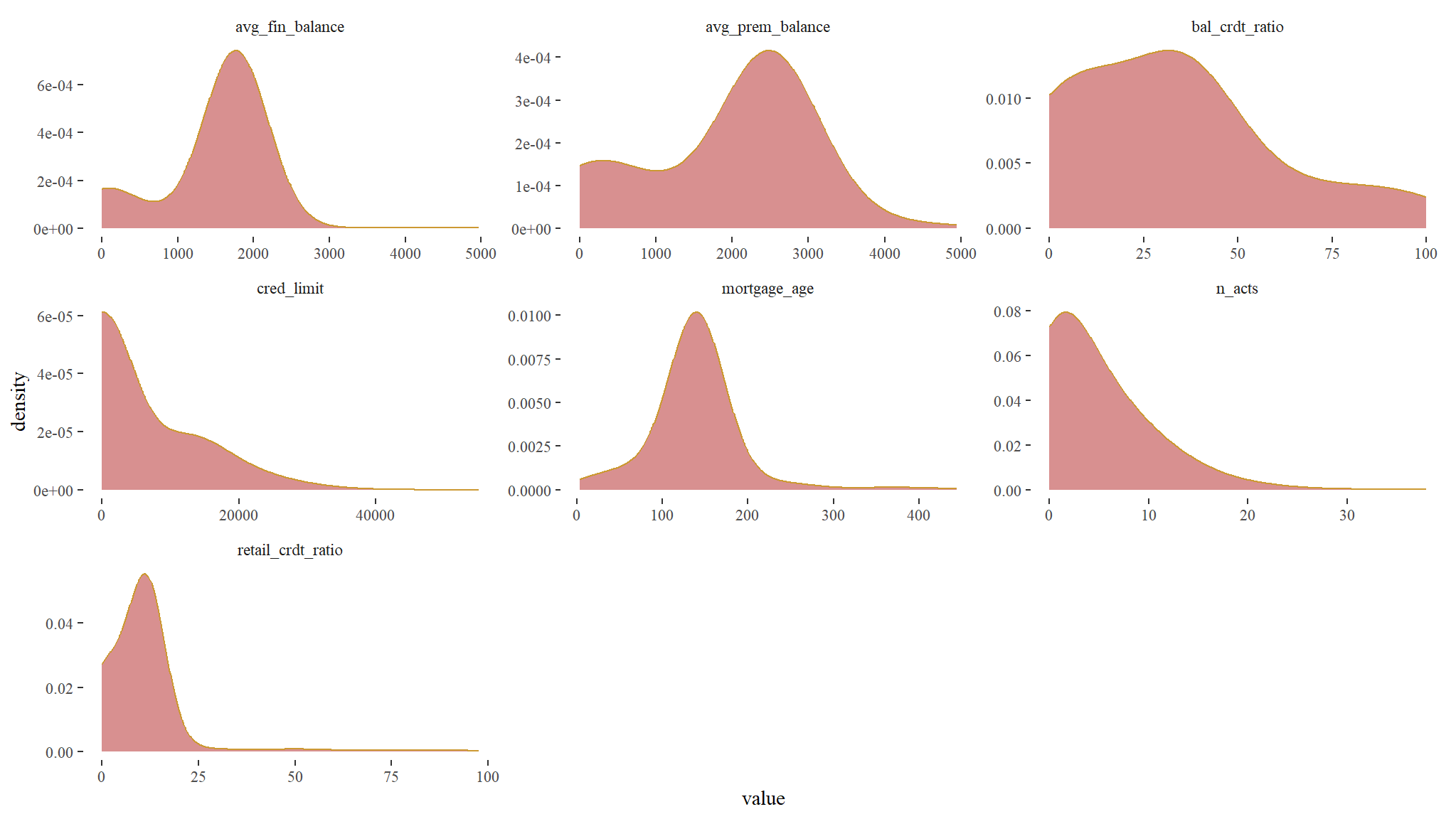
correlation
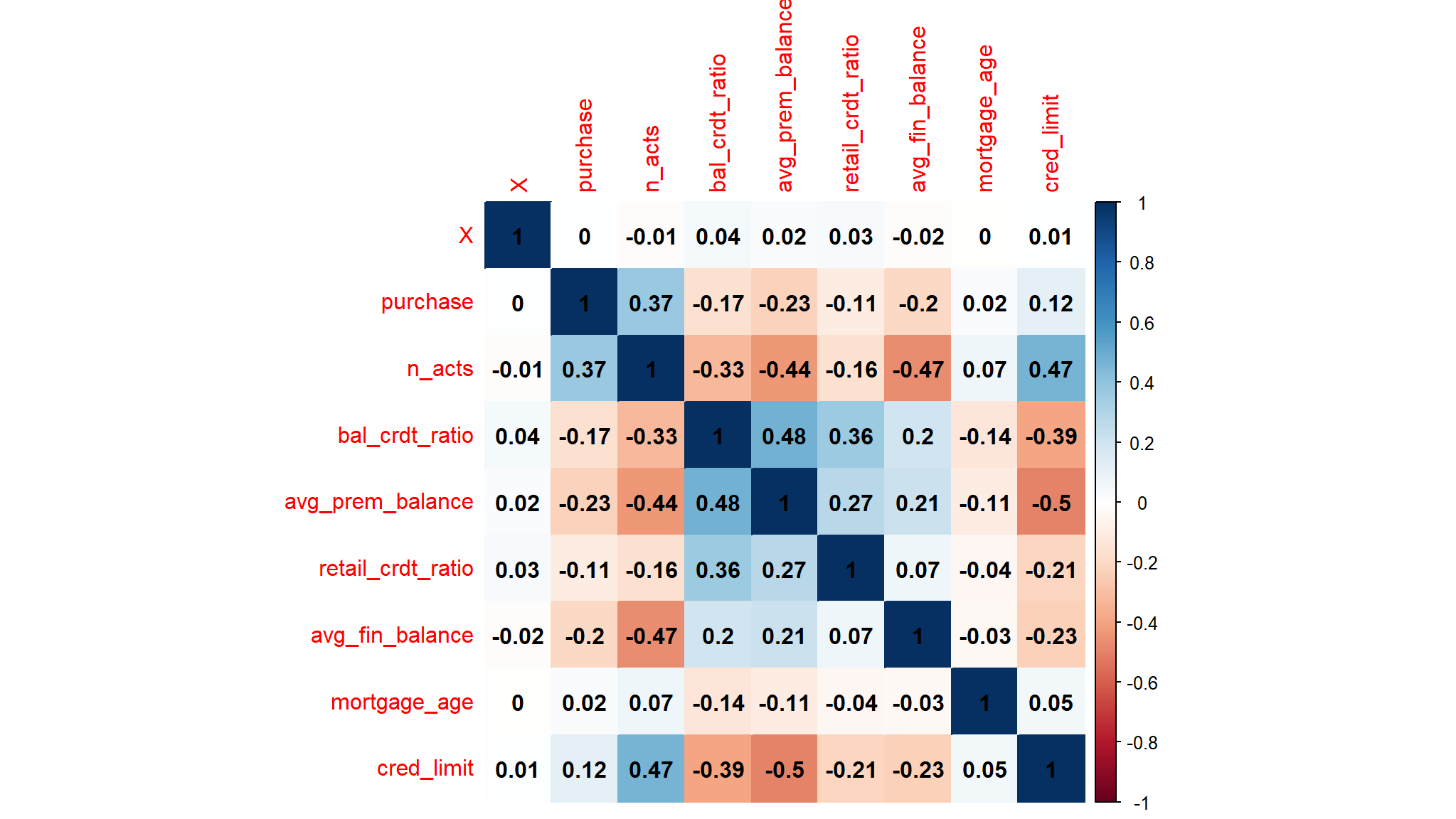
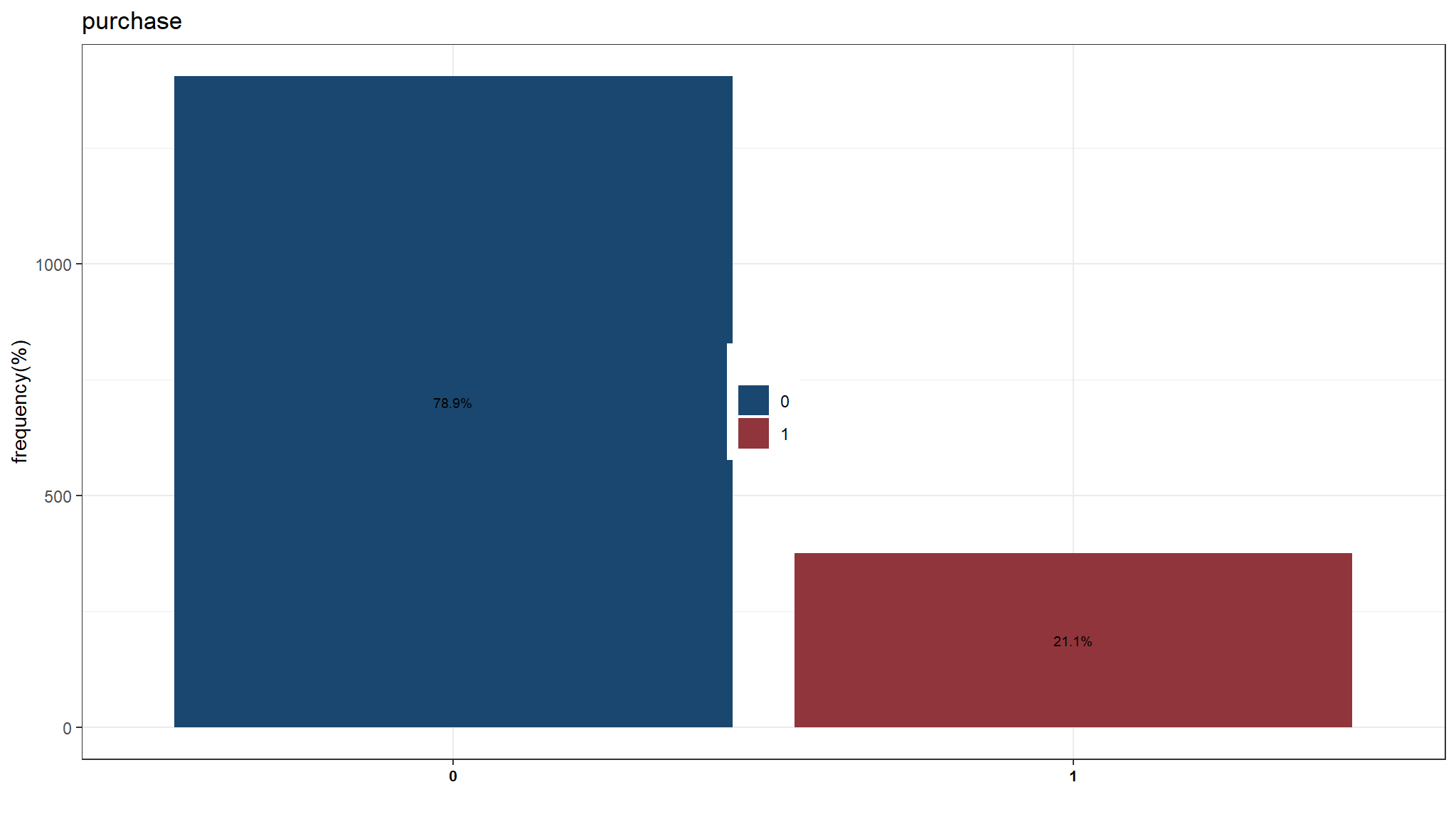
Initial model development
GLM logistic
# Fit a logistic model
log_mod_glm <- glm(purchase ~ mortgage_age, data = csale,
family = binomial)
summary(log_mod_glm)
#>
#> Call:
#> glm(formula = purchase ~ mortgage_age, family = binomial, data = csale)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -1.46292 0.16659 -8.781 <2e-16 ***
#> mortgage_age 0.00103 0.00112 0.919 0.358
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 1832.4 on 1778 degrees of freedom
#> Residual deviance: 1831.6 on 1777 degrees of freedom
#> AIC: 1835.6
#>
#> Number of Fisher Scoring iterations: 4AIC(log_mod_glm)
#> [1] 1835.554GAM logistic
# Fit a logistic model
log_mod_gam <- gam(purchase ~ s(mortgage_age), data = csale,
family = binomial,
method = "REML")
summary(log_mod_gam)
#>
#> Family: binomial
#> Link function: logit
#>
#> Formula:
#> purchase ~ s(mortgage_age)
#>
#> Parametric coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -1.34131 0.05908 -22.7 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Approximate significance of smooth terms:
#> edf Ref.df Chi.sq p-value
#> s(mortgage_age) 5.226 6.317 30.02 4.11e-05 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> R-sq.(adj) = 0.0182 Deviance explained = 1.9%
#> -REML = 910.49 Scale est. = 1 n = 1779AIC(log_mod_gam)
#> [1] 1811.976anova(log_mod_glm,log_mod_gam,test="LRT")
#> Analysis of Deviance Table
#>
#> Model 1: purchase ~ mortgage_age
#> Model 2: purchase ~ s(mortgage_age)
#> Resid. Df Resid. Dev Df Deviance Pr(>Chi)
#> 1 1777.0 1831.5
#> 2 1772.8 1797.6 4.2261 33.963 9.982e-07 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- the AIC already suggests that a logistic model under a
GAMis better than that in aGLM - The anova test also suggests that a model with a smooth term performs better than the one without
fit a full model
GAM logistic
# Fit a logistic model
log_mod_gam2 <- gam(purchase ~ s(n_acts) + s(bal_crdt_ratio) +
s(avg_prem_balance) + s(retail_crdt_ratio) +
s(avg_fin_balance) + s(mortgage_age) +
s(cred_limit),
data = csale,
family = binomial,
method = "REML")
# View the summary
summary(log_mod_gam2)
#>
#> Family: binomial
#> Link function: logit
#>
#> Formula:
#> purchase ~ s(n_acts) + s(bal_crdt_ratio) + s(avg_prem_balance) +
#> s(retail_crdt_ratio) + s(avg_fin_balance) + s(mortgage_age) +
#> s(cred_limit)
#>
#> Parametric coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -1.64060 0.07557 -21.71 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Approximate significance of smooth terms:
#> edf Ref.df Chi.sq p-value
#> s(n_acts) 3.474 4.310 93.670 < 2e-16 ***
#> s(bal_crdt_ratio) 4.308 5.257 18.386 0.00319 **
#> s(avg_prem_balance) 2.275 2.816 7.800 0.04889 *
#> s(retail_crdt_ratio) 1.001 1.001 1.422 0.23343
#> s(avg_fin_balance) 1.850 2.202 2.506 0.27889
#> s(mortgage_age) 4.669 5.710 9.656 0.13356
#> s(cred_limit) 1.001 1.002 23.066 2.37e-06 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> R-sq.(adj) = 0.184 Deviance explained = 18.4%
#> -REML = 781.37 Scale est. = 1 n = 1779AIC(log_mod_gam2)
#> [1] 1542.598
# Plot on the log-odds scale
plot(log_mod_gam2, pages = 1)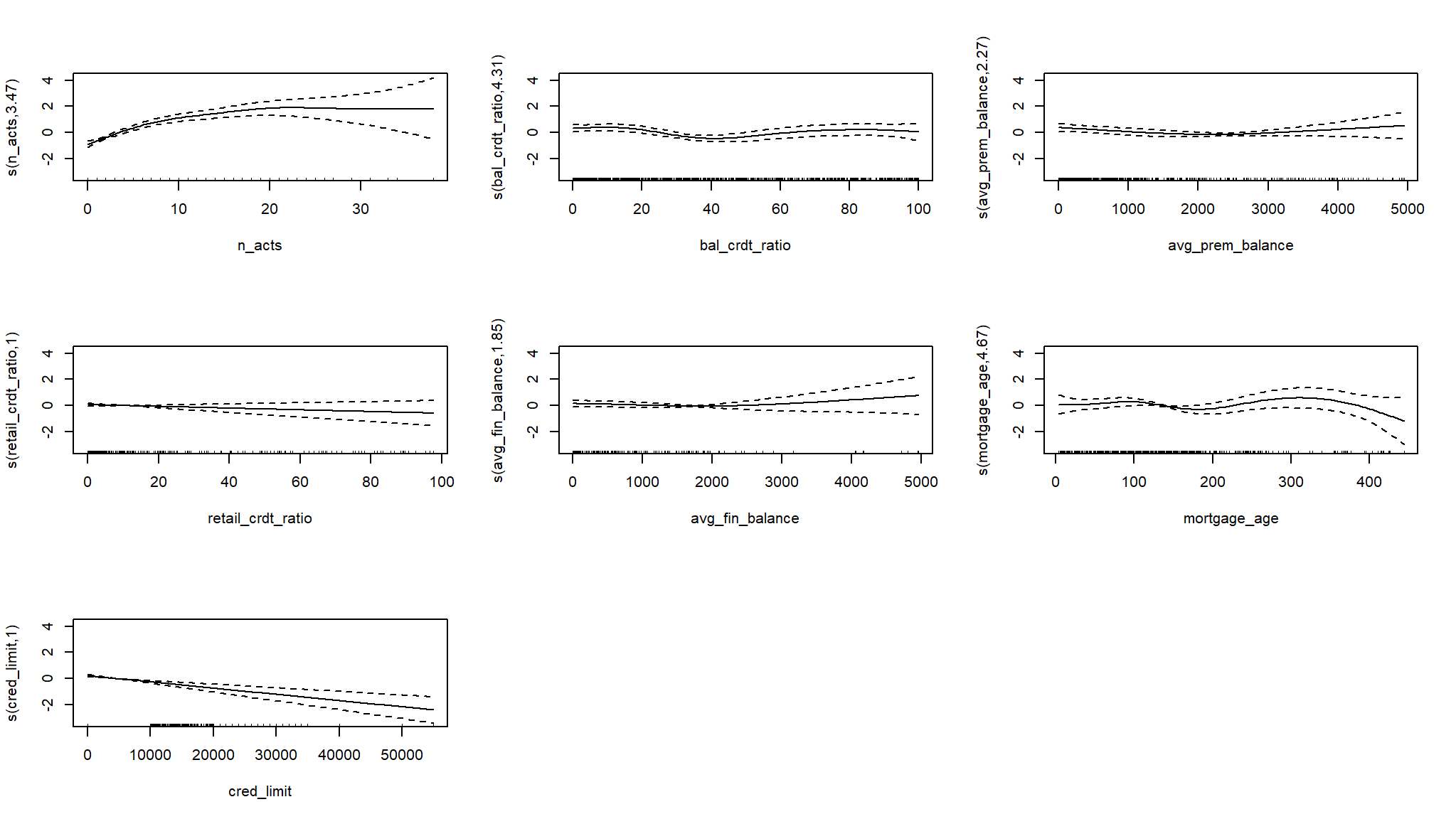
# Plot on the probability scale
plot(log_mod_gam2, pages = 1, trans = plogis)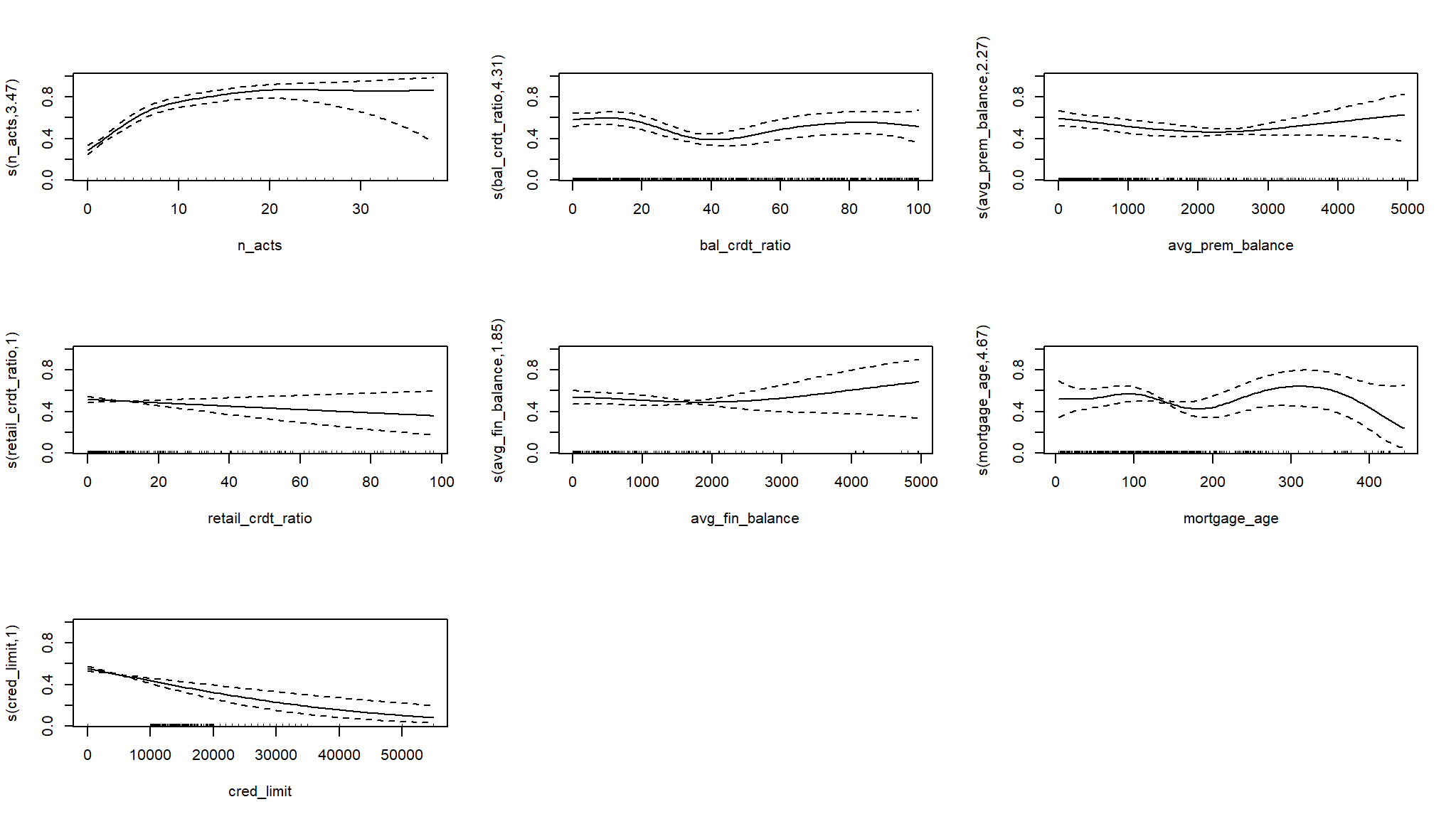
# Plot with the intercept
plot(log_mod_gam2, pages = 1, trans = plogis,
shift = coef(log_mod_gam2)[1])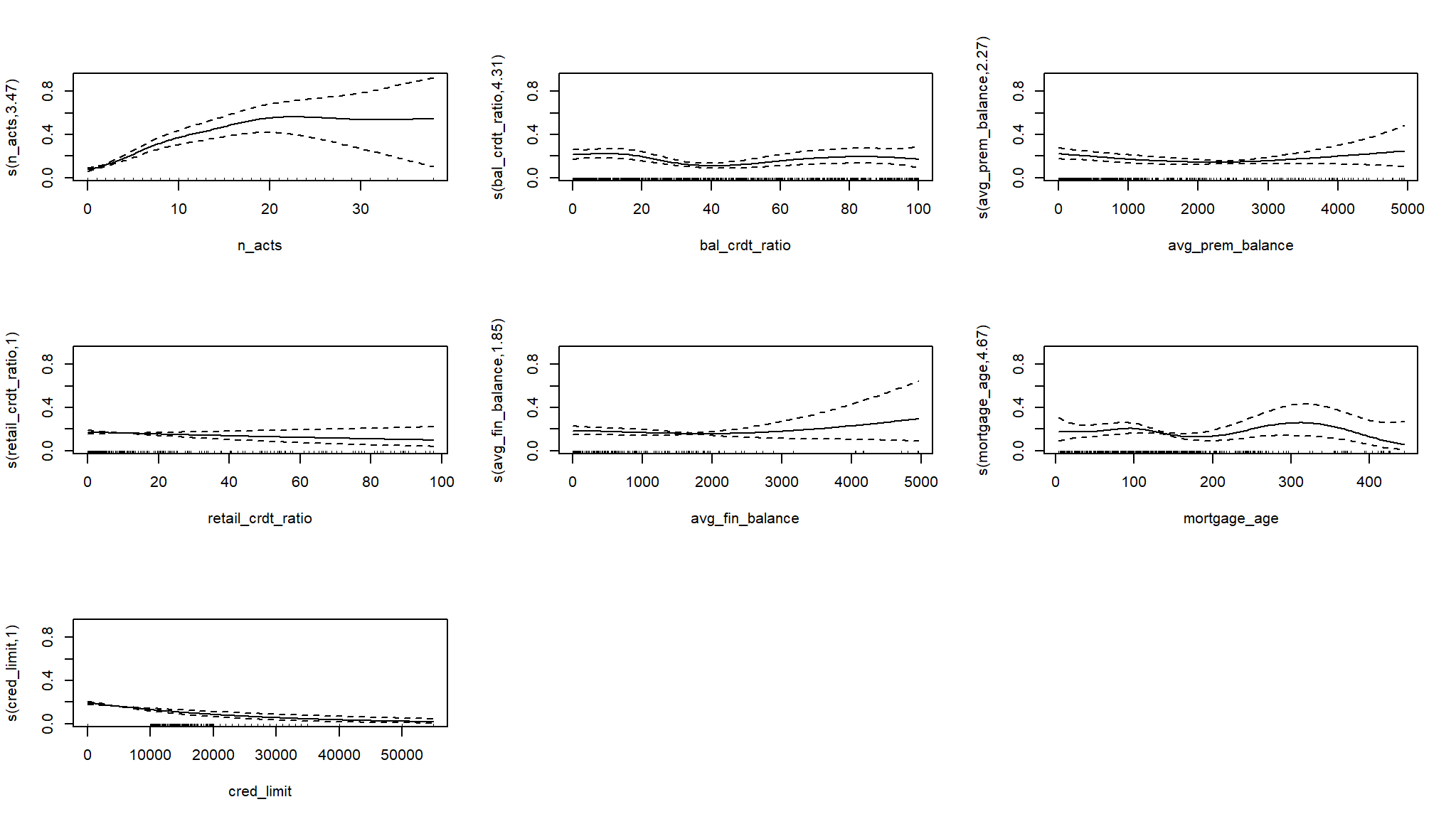
# Plot with intercept uncertainty
plot(log_mod_gam2, pages = 1, trans = plogis,
shift = coef(log_mod_gam2)[1], seWithMean = TRUE,
rug = FALSE, shade = TRUE, shade.col = "lightgreen" , col = "purple")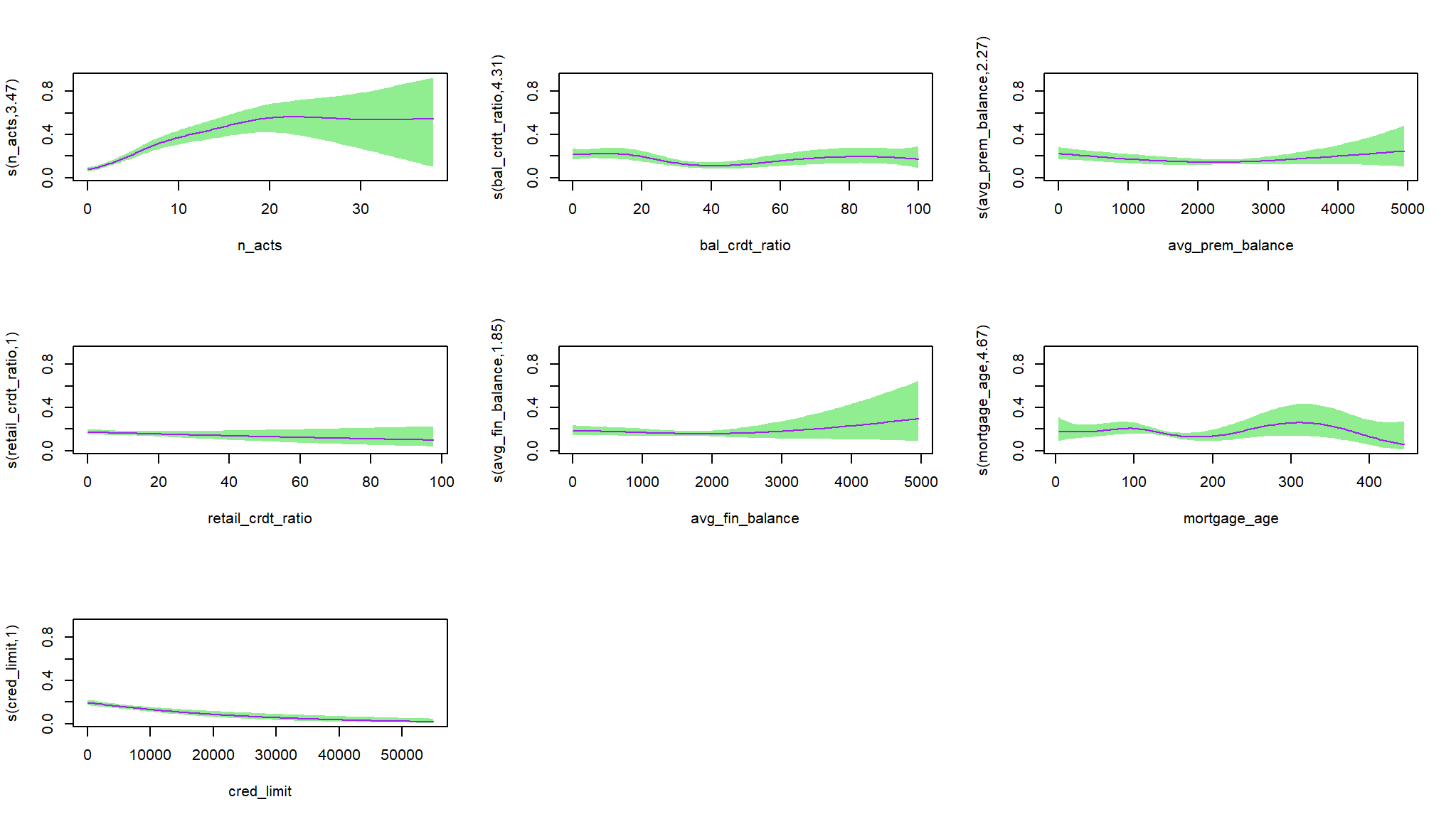
GLM logistic
log_m <- glm(purchase ~ n_acts + bal_crdt_ratio +
avg_prem_balance + retail_crdt_ratio +
avg_fin_balance + mortgage_age +
cred_limit,
data = csale,
family = binomial)
summary(log_m)
#>
#> Call:
#> glm(formula = purchase ~ n_acts + bal_crdt_ratio + avg_prem_balance +
#> retail_crdt_ratio + avg_fin_balance + mortgage_age + cred_limit,
#> family = binomial, data = csale)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -1.052e+00 2.901e-01 -3.628 0.000285 ***
#> n_acts 1.483e-01 1.467e-02 10.109 < 2e-16 ***
#> bal_crdt_ratio -4.485e-03 3.003e-03 -1.493 0.135310
#> avg_prem_balance -2.335e-04 7.006e-05 -3.333 0.000860 ***
#> retail_crdt_ratio -6.247e-03 5.916e-03 -1.056 0.291052
#> avg_fin_balance -1.157e-04 9.104e-05 -1.271 0.203606
#> mortgage_age -8.033e-04 1.134e-03 -0.709 0.478596
#> cred_limit -3.718e-05 9.579e-06 -3.881 0.000104 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 1832.4 on 1778 degrees of freedom
#> Residual deviance: 1580.0 on 1771 degrees of freedom
#> AIC: 1596
#>
#> Number of Fisher Scoring iterations: 4AIC(log_m)
#> [1] 1596.011- again the GAM logistic performs better with a much smaller
AIC
Lets see how these compare on new predictions
set up data for modeling
set up a recipe
train_rcp <- recipe(purchase ~ ., data=csale)
# Prep and bake the recipe so we can view this as a seperate data frame
training_df <- train_rcp |>
prep() |>
juice()Set up engines for different models
initialise a GLM model
log_mod_glm <- logistic_reg() |>
set_engine('glm') |>
set_mode("classification")add to workflow
lr_wf <-
workflow() |>
add_model(log_mod_glm) |>
add_recipe(train_rcp)lr_fit <-
lr_wf |>
fit(data=train)Extracting the fitted data
I want to pull the data fits into a tibble I can explore. This can be done below:
lr_fitted <- lr_fit |>
extract_fit_parsnip() |>
tidy()I will visualise this via a bar chart to observe my significant features:
Use fitted logistic regression model to predict on test set
We are going to now append our predictions from our model we created as a baseline to append to the predictions we already have in the predictions data frame:
testing_lr_fit_probs <- predict(lr_fit, test, type='prob')
testing_lr_fit_class <- predict(lr_fit, test)
predictions<- cbind(test, testing_lr_fit_probs, testing_lr_fit_class)
predictions <- predictions |>
dplyr::mutate(log_reg_model_pred=.pred_class,
log_reg_model_prob=.pred_1) |>
dplyr::select(everything(), -c(.pred_class, .pred_0)) |>
dplyr::mutate(log_reg_class_custom = ifelse(log_reg_model_prob >0.7,"Yes","No")) |>
dplyr::select(-.pred_1)Confusion matrix
predictions |>
conf_mat(purchase,log_reg_model_pred)
#> Truth
#> Prediction 0 1
#> 0 340 72
#> 1 11 22predictions |>
conf_mat(purchase,log_reg_model_pred) |>
autoplot(type="heatmap")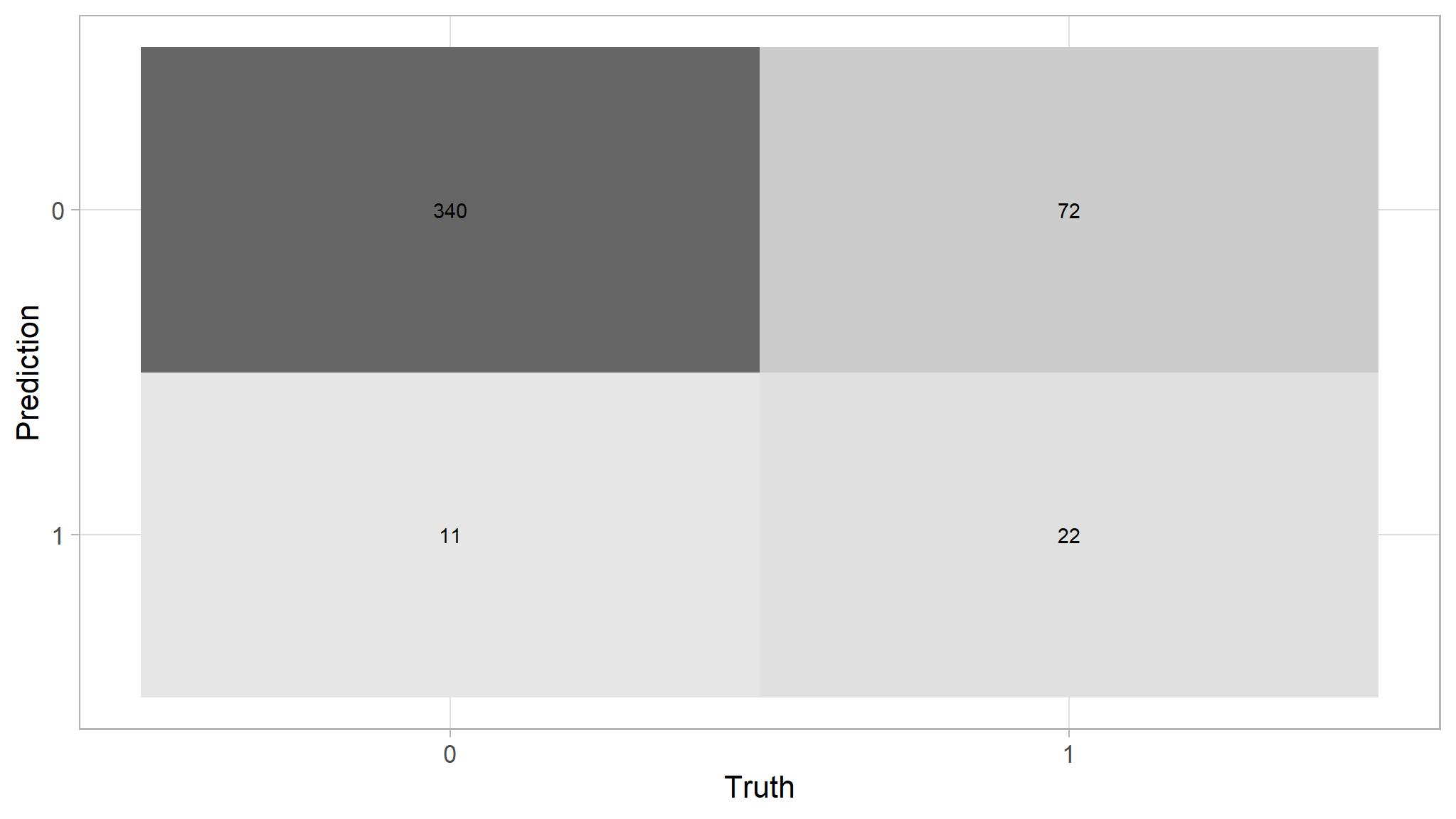
initialise a GAM model
log_mod_gam <- gen_additive_mod() |>
set_engine('mgcv',method="REML") |>
set_mode("classification")add to workflow
Extracting the fitted data
I want to pull the data fits into a tibble I can explore. This can be done below:
lr_fitted <- lr_fit |>
tidy()I will visualise this via a bar chart to observe my significant features:
Use fitted logistic regression model to predict on test set
We are going to now append our predictions from our model we created as a baseline to append to the predictions we already have in the predictions data frame:
predictions |>
conf_mat(purchase,log_reg_model_pred)
#> Truth
#> Prediction 0 1
#> 0 335 72
#> 1 16 22predictions |>
conf_mat(purchase,log_reg_model_pred) |>
autoplot(type="heatmap")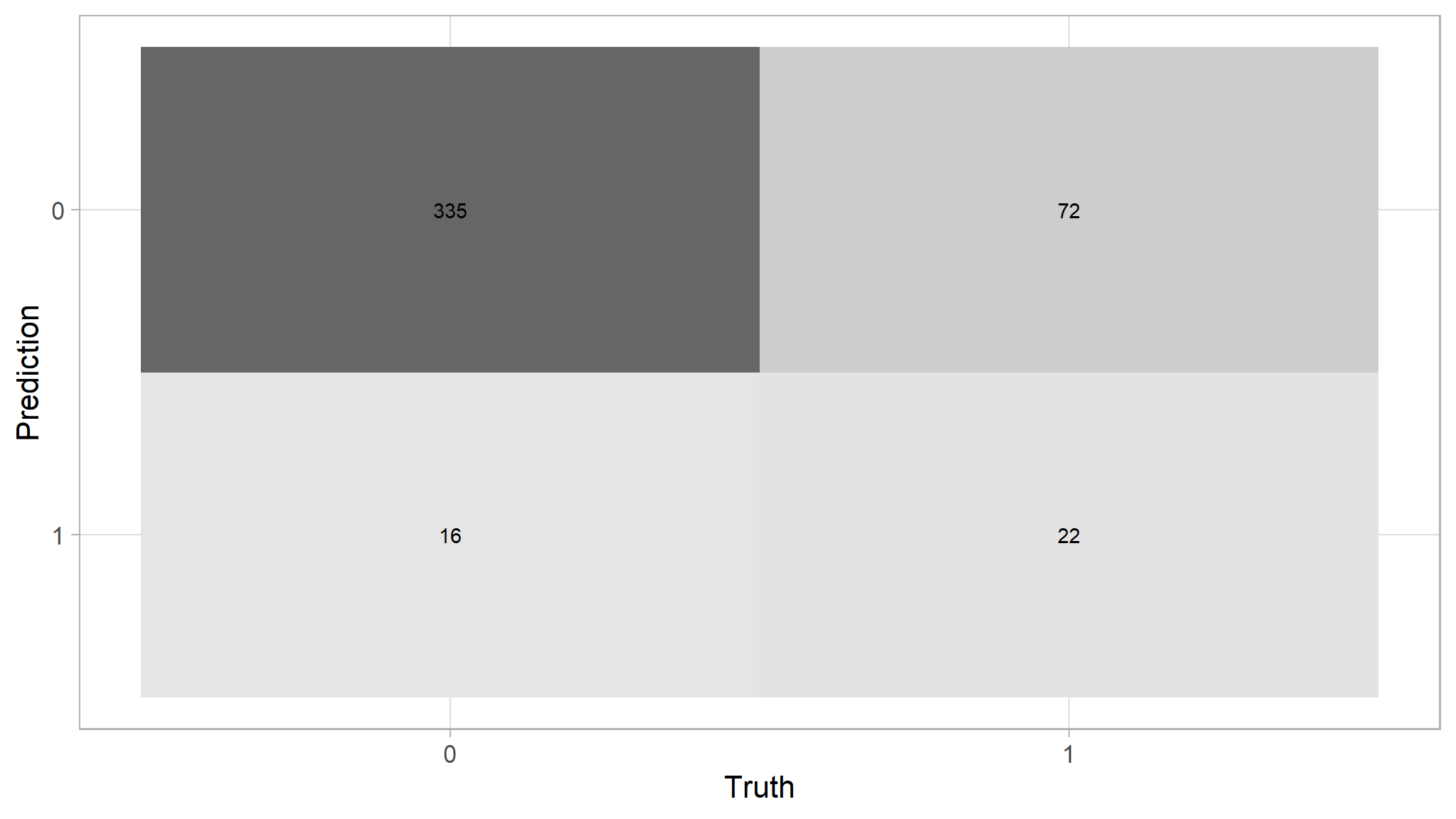
- The generalised Additive logistic regression performs a bit poor than the glm
- the difference is just one percent accuracy